Polymerase chain reaction (PCR) is a widely-used technique in molecular biology. Thanks to the COVID-19 pandemic, the general public has become far more aware of PCR and its use in molecular diagnostics.
In this blog, we will describe the information that can be gleaned from ‘multiplex’ PCR, and how this information can be used to diagnose infectious diseases and select appropriate treatments.
Infectious Diseases
Infectious Diseases: a short introduction
Bacterial and viral infections are associated with numerous medical conditions. Infectious diseases range from relatively mild illnesses (such as chicken pox in infants) to diseases with staggeringly high mortality rates (such as Ebola haemorrhagic fever).
Even the ‘common cold’ is a viral infection, often caused by rhinoviruses or coronaviruses.
How big a problem are infectious diseases?
The recent COVID-19 pandemic led to a worldwide focus on infectious diseases. As of March 2023, COVID-19 has been linked to 150,000 hospital deaths in England alone [1]. Furthermore, the World Health Organisation (WHO) estimates that human immunodeficiency virus (HIV; the causative agent of AIDS), viral hepatitis and sexually transmitted infections (STIs) collectively cause over two million deaths per year [2].
These examples represent the tip of the iceberg, as there are many other infectious diseases afflicting human populations worldwide. A fast and accurate diagnosis – and an appropriate medical intervention – are key to relieving the burden of infectious disease across the world.
Multiplex PCR
What is PCR?
Since its invention by Kary Mullis in 1985, PCR has become a vital tool for molecular biologists [3]. In a typical PCR, a DNA sequence is amplified using two short oligonucleotide molecules (referred to as ‘primers’) which anneal to specific sites at either side of the target sequence.
Amplification of the DNA sequence between the two primer binding sites is then achieved via thermal cycling. As PCR progresses, a small number of copies of the original DNA sequence are amplified to generate many millions of copies. This allows specific DNA & RNA sequences to be detected within complex mixtures.
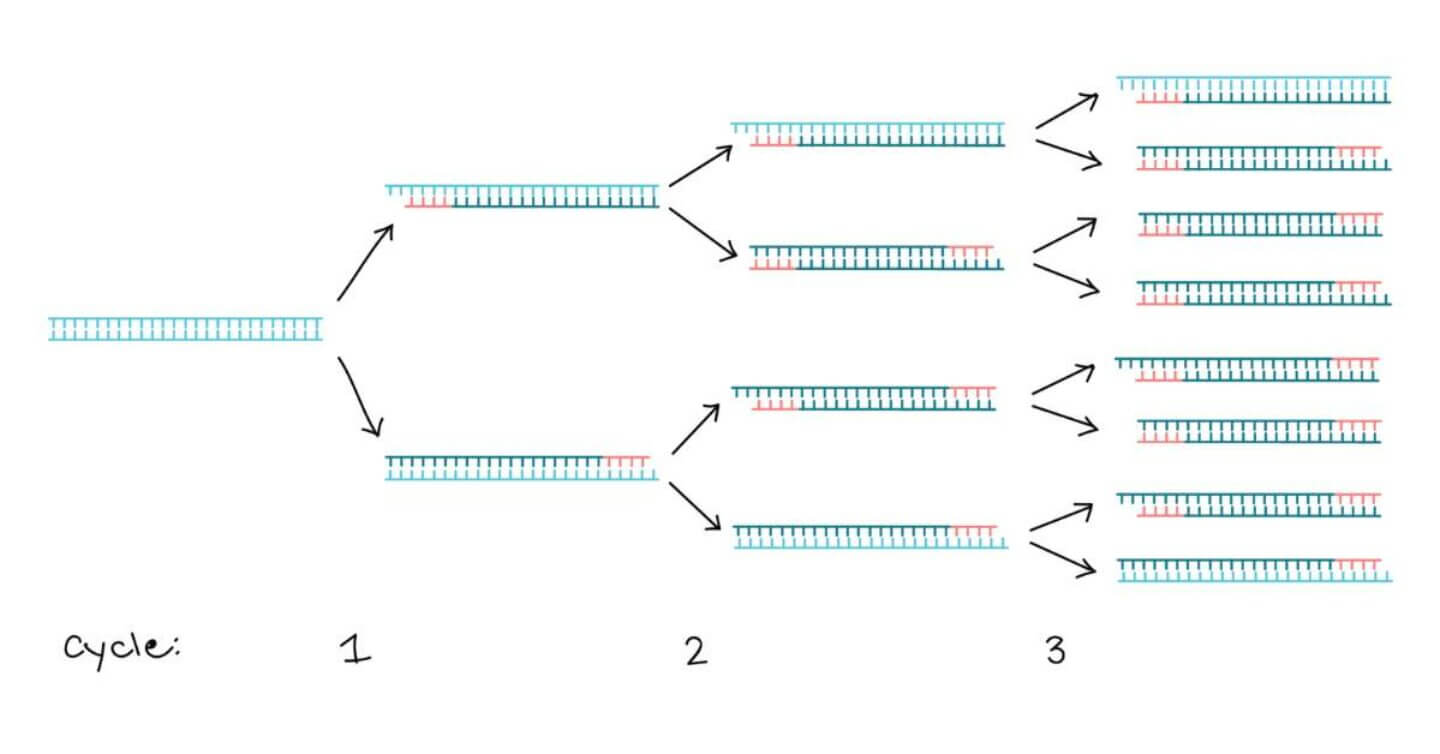
What is multiplex PCR?
As outlined above, a ‘basic’ PCR mixture includes just one pair of oligonucleotide primers. In contrast, a ‘multiplex’ PCR incorporates multiple primer pairs into a single mixture. This means that it is possible to amplify many different DNA sequences at the same time.
Importantly for us, multiplex PCR is a fast, accurate and specific technique for detecting infectious diseases in biological specimens [4].
Using Multiplex PCR to diagnose Infectious Diseases
Multiplex PCR: an ideal diagnostic tool
The extraction of genetic material from bacteria or viruses in a clinical specimen can provide template DNA for PCR. The amplification of a pathogen-specific DNA sequence can be interpreted as a ‘Positive’ result for an infectious disease. In recent years, multiplex PCR has expanded this simple paradigm.
The chances of appropriate treatment are increased if an infection is diagnosed correctly. However, many different infectious diseases can cause patients to present similar clinical symptoms, often resulting in misdiagnosis.
As we know, infection with SARS-CoV-2 (the causative agent of COVID-19) can lead to coughing, fever, breathlessness and fatigue in many patients. Unfortunately for clinicians, similar symptoms can also be caused by influenza (‘Flu’) and respiratory syncytial virus (RSV).
Similarly, STIs such as chlamydia, gonorrhoea and syphilis can be easily mistaken for each other, even by clinicians.
To address these challenges, multiplex PCR mixes can be developed to include primer pairs targeting DNA sequences in many different bacteria or viruses.
For example, a patient may present symptoms that are consistent with a respiratory tract infection, such as COVID-19 or Flu. The patient would be asked to provide a clinical specimen, such as a throat swab. This specimen would then be analysed using a multiplex PCR assay. If a SARS-CoV-2-specific sequence was amplified, but an influenza-specific sequence was NOT amplified, the patient would be diagnosed with COVID-19, not Flu.
In addition, the emergence of antibiotic resistance in bacteria means that a particular therapeutic intervention may be effective for some patients but ineffective in others. Antibiotic resistance is caused by ‘point mutations’ at key areas of the bacterial genome.
Again, multiplex PCR is an ideal diagnostic tool to address this problem. By using genome sequence data, it is possible to design primers that will only bind to the ‘normal’ bacterial genome, and other primers that will only bind when point mutations are present. By distinguishing these PCR products, it is possible to diagnose an infection AND to select an effective antibiotic.

Are there any other advantages of multiplex PCR?
When produced at scale, multiplex PCR assays can be a relatively inexpensive and rapid tool for healthcare systems to use.
Many traditional diagnostic techniques require bacterial cells to be grown on plates, meaning that it can take days for results to be available. In contrast, multiplex PCR assay results can be returned in under an hour.
Furthermore, it is possible to run a large number of multiplex PCR assays simultaneously, on the same lab-based instrument. This allows many clinical specimens to be analysed at once with a high throughput.
Another advantage of PCR-based diagnostics is the high sensitivity that they provide, with the amplification & detection of a single DNA molecule being theoretically possible! This means that a patient can be diagnosed in the very early stages of an infection; sometimes before symptoms present themselves.
As a result, the patient can be managed appropriately. The patient can also isolate themselves to ensure that they do not pass on an infection to their friends, family or work colleagues.
What about limitations of multiplex PCR?
Although these are many advantages of multiplex PCR, there are also disadvantages.
Multiplex PCR assays usually require expensive thermal cyclers and centralised laboratory infrastructure, as well as skilled users to prepare the reactions and interpret the results. This is not the case for other diagnostic tools such as lateral flow tests (LFTs), which can be used by patients at the point of care.
In addition, the high mutation rates in bacterial and viral genomes can affect the binding of PCR primers to their target sites. This can render PCR assays ineffective when attempting to diagnose new variants.
Another disadvantage is the time required to extract genetic material from a clinical specimen prior to PCR analysis. This is often an essential step, as biological contaminants can inhibit PCR, leading to ‘false-negative’ results being returned.
QuantuMDx and the fight against Infectious Diseases
Since the company was founded in 2008, QuantuMDx has focussed on providing high quality, affordable, rapid molecular diagnostics to patients at the point of need.
Multiplex PCR is a key element of QMDx technology. The Q-POC™ SARS-CoV-2, Flu A/B & RSV Assay is now commercially available and delivers rapid results in approximately 35 minutes.
In this assay, a microfluidic multiplex PCR panel assay allows SARS-CoV-2, Flu A, Flu B and RSV infections to be detected and distinguished without the need for manual processing of a clinical specimen. Results are provided to the user via a simple interface, without the need for any clinical expertise or data interpretation.
QuantuMDx is currently expanding its product portfolio to include many more molecular diagnostic tests.
References
[1] https://www.england.nhs.uk/statistics/statistical-work-areas/covid-19-deaths/, NHS England, 2023.
[2] W. H. Organisation, “Global health sector strategies on, respectively, HIV, viral hepatitis and sexually transmitted infections for the Period 2022-30,” 2022.
[3] https://www.ncbi.nlm.nih.gov/probe/docs/techpcr/, NCBI.
[4] S. Yang and R. Rothman, “PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings.,” Lancet Infect Dis, vol. 4, no. 6, pp. 337-48, 2004.
[5] N. Sea-Liang, A. Sereemaspun, K. Patarakul, J. Gaywee, W. Rodkvamtook, N. Srisawat, S. Wacharaplusadee and T. Hemachudha, “Development of multiplex PCR for neglected infectious diseases,” PLoS Negl Trop Dis, vol. 13, no. 7, pp. 1-12, 2019.








