Multiplex PCR has become a mainstream method to rapidly diagnose infectious diseases. In this short post, we introduce an emerging branch of molecular diagnostic technology: the detection of mRNA signatures.
We tackle the following questions:
- What is mRNA?
- What is an mRNA signature?
- Why are mRNA signatures of interest?
- How can mRNA signatures be applied to infectious disease diagnostics?
QuantuMDx have adapted a well-established mRNA signature for tuberculosis (TB) for use as a multiplex PCR test and we discuss how this could be a vital tool for the future of TB testing.
What is mRNA?
First things first: what is mRNA and how does it differ to DNA?
DNA (full name DeoxyriboNucleic Acid) is a double-stranded molecule structured in a double helix formation that is bundled up in a cell structure called the nucleus. DNA carries information (genes and regulatory instructions) that is necessary for any living thing to exist.
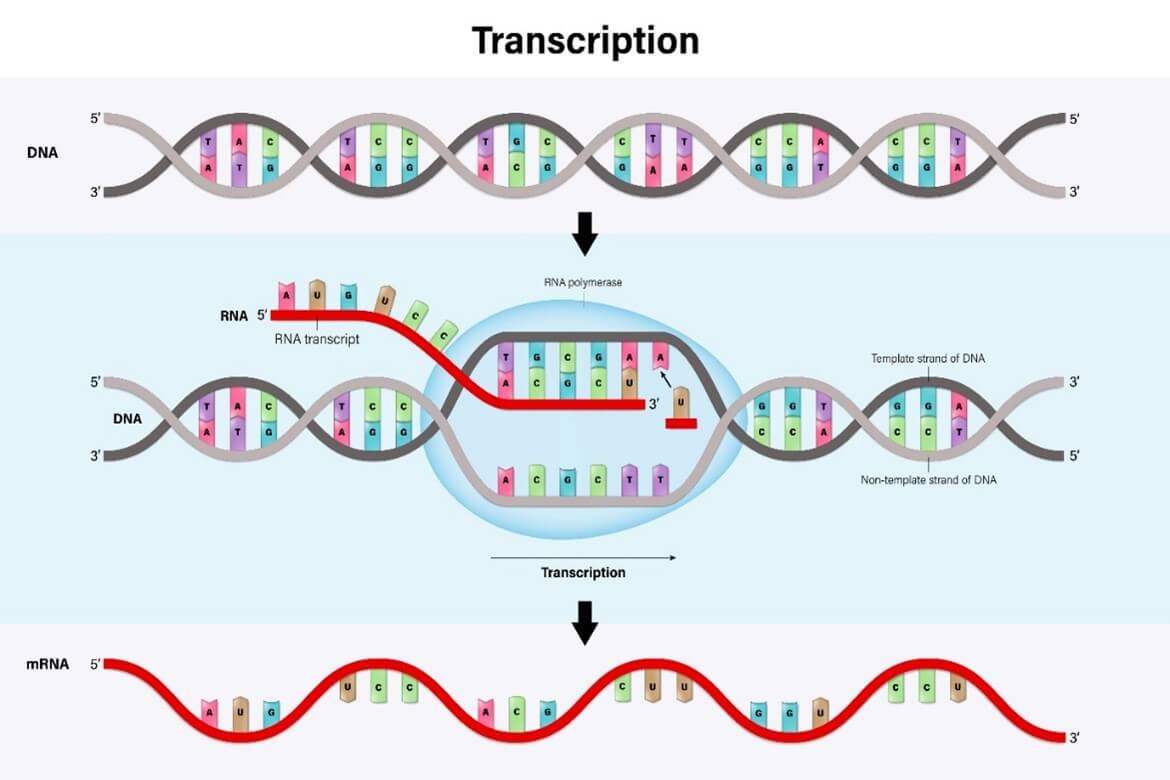
In order for the DNA genetic code to be translated into proteins, the building blocks of physical cells, the DNA double helix is separated into single strands so that it can be copied by polymerase enzymes (Fig. 1A).
This copying process, known as transcription, generates a single strand of RNA (RiboNucleic Acid) called messenger RNA (or mRNA) that is a mirror image (or ‘complementary’) to the DNA strand (Fig. 1A).
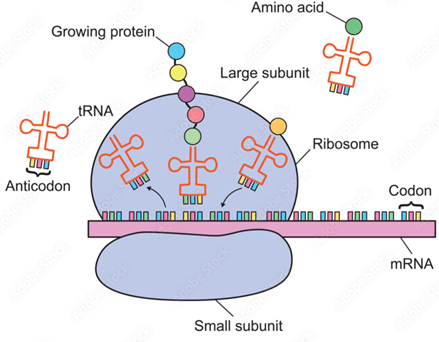
The mRNA moves to the ribosome, where the strict order of codons (the name for three consecutive nucleotides) is used to recruit the correct amino acids in the correct order to synthesise a specific protein. Thus, the DNA code is ‘translated’ into proteins (Fig. 1B).
Why is the difference between DNA and mRNA important to understand?
First, consider that replicating all genes encoded by DNA all of the time would require lots of energy and would be very inefficient.
It would also make it difficult to change the abundance of particular proteins at different times, which is crucial for all basic cellular functions and for a cell to respond to changes in its environment.
Instead, only genes that are needed in particular cells at particular times are switched on (‘expressed’) by producing mRNA.
So, now we understand the function of mRNA, let’s explore the concept of mRNA signatures.
What is an mRNA signature?
An mRNA transcript is the result of a gene that is being expressed at a given time.
Adding ‘ome’ to the end of certain words has become more commonplace and broadly speaking means ‘mass’ or ‘complete set’. For instance, genome describes all of the genes of an organism, and gut microbiome describes all of the microorganisms in your gut.
Therefore, ‘transcriptome’ refers to all the mRNA transcripts in a cell, which in turn represents all the genes that are being actively expressed in a cell at any one time.
As you can imagine, the amount of information associated with the transcriptome of a cell is enormous and cannot practicably be used in a diagnostic capacity.
An mRNA ‘signature’ is a much smaller, specific group of mRNA transcripts that are uniquely characteristic of a physical (phenotypic) state, such as a disease or dysfunctional state. We now know that these signatures can potentially be used for diagnostic purposes.
A lengthy process of highly detailed analysis is required to identify ‘differentially expressed genes’ that are associated with the particular clinical condition of interest. The expression of some genes may be increased (‘upregulated’) while others are decreased (‘downregulated’).
The extent of any upregulation or downregulation may also be highly important. Once a putative mRNA signature has been identified, extensive validation is required to ensure that the transcripts are only associated with the disease or dysfunction and are not expressed in healthy situations.
Why are mRNA signatures of interest?
Detection of active gene expression via an mRNA signature offers advantages over ‘traditional’ PCR-based diagnostics.
Multiplex PCR methods largely rely on detection of the presence or absence of genes associated with a particular disease or infection. The detection of mRNA transcripts can provide extra information by telling us if these genes are being upregulated (switched on) or downregulated (switched off), and how these transcripts may be functioning in relation to others.
Quantifying mRNA transcripts represents a much more dynamic way to investigate cellular processes in the body (host responses) that may be occurring at a particular time in response to a dysfunctional state, disease or infection. For instance, detection of mRNA signatures may be used to detect reactivation of disease or response to treatment.
mRNA signatures are of interest as the basis of diagnostic tests because they lend themselves to blood-based testing. If testing can be completed from a whole blood sample (especially one obtained by a fingerstick sample), testing becomes more patient-friendly and more accessible compared with collecting other sample types, which may be less easy to obtain, such as a biopsy sample for cancer diagnosis or venous blood sample for diagnosis of sepsis.
As with any new technology in early stages of clinical development, there are certain hurdles to be overcome before mRNA-based diagnostics can be implemented into clinical practice. Challenges that are keeping researchers busy include (a) identification of appropriate control groups for clinical trials, (b) acquiring appropriate reference tests for comparison, and (c) how to present complex results in an easy-to-interpret way.
How can mRNA signatures be applied to infectious disease diagnostics?
An mRNA signature may act as biomarker (i.e. a biological indicator of a particular disease) and as such are being explored for a wide array of health conditions, such as identification of certain cancers (Čelešnik & Potočnik, 2023; Jiang et al., 2022), prediction of kidney transplant rejection (Fekih et al., 2021), and understanding autoimmune diseases (Nagafuchi et al., 2022).
In terms of infectious disease diagnostics, mRNA signatures are being developed to discriminate between bacterial and viral infections (Bodkin et al., 2022), detect and predict the progression of life-threatening infections such as sepsis (Lukaszewski et al., 2022; Baghela et al., 2022), and offer advances for the detection of tuberculosis (Mulenga et al., 2020).
Use of an mRNA-signature for tuberculosis detection
At QuantuMDx, we have developed a multiplex PCR to detect host-response mRNA transcripts present in whole blood that are associated with tuberculosis.
Based upon an existing 6-gene mRNA signature (RISK6), this laboratory-based multiplex one-step RT-qPCR assay, known as the TB-Host Immune Response Assay (TB-HIRA), simultaneously amplifies and detects six mRNA transcripts directly from extracted RNA in a single reaction on a benchtop thermocycler.
Currently available for research use only, once fully validated the laboratory-based multiplex aims to provide a non-sputum-based testing solution for tuberculosis.
We are developing the TB-HIRA laboratory-based test for use on our point-of-care platform, the Q-POC™. In addition to direct sample-to-result testing, the Q-POC aims to automatically generate a result in ~30 minutes with a small volume of whole blood (µL) from a fingerstick sample.
We encourage you to get in touch if you would like to evaluate the QuantuMDx laboratory-based assay in your research or be an early adopter of the TB-HIRA on the Q-POC™.
Figure 1. Diagrammatic representation of the process of transcription of a double-stranded DNA molecule to a single-stranded RNA molecule [A] and how mRNA is translated into proteins during protein synthesis [B].
Images courtesy of Adobe Stock: [A] 476622668 and [B] 500809247.








